A research article recently published in the Nature Communications journal presents an algorithm inspired from computer vision that can detect patterns in contact maps. The paper called "Computer vision for pattern detection in chromosome contact maps" outlines the development of the Chromosight algorithm which provides insights into genomes by using visible patterns on genome-wide contact maps generated by chromosome conformation capture approaches such as Hi-C.
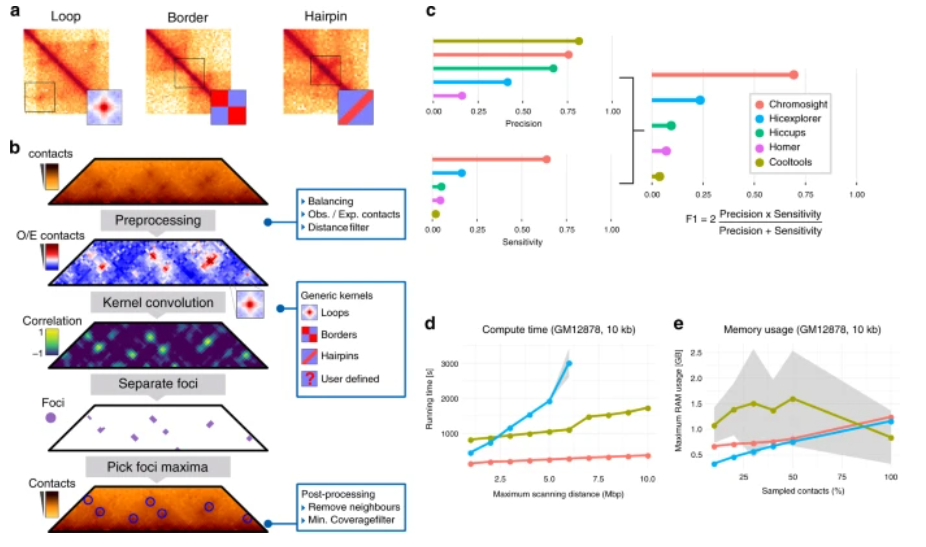
While most tools aiming at detecting DNA loops in contact maps rely on computationally intensive approaches and are developed mostly from, and for, human data, Chromosight is an algorithm that, when applied on mammalian, bacterial, viral and yeast genome-wide contact maps, quickly and efficiently detects and quantifies any type of pattern, with a specific focus on chromosomal loops.
Authors of the article, amongst whom IGNITE researchers, performed a number of tests of the Chromosight tool in comparison with other tools used for the same purpose. They outline the Chromosight algorithm’s comparative advantage to similar tools as it does not require any prior training dataset and works well with default parameters on data generated with various protocols.
Read the article here.