IGNITE research uses genomic comparisons to identify considerable differences in the general structure of the genome and the makeup and history of various gene families of biological relevance to habitat adaptation.
This study uses a comparative genomics approach for a deeper appreciation of the structure and causes of the deeply diverging lineages in the Placozoa. Placozoans are a phylum of tiny (approximately 1 mm) marine animals that are found worldwide in temperate and tropical waters. They are characterized by morphological simplicity, with only a handful of cell types, no neurons, no tissue organization, and even no axial polarity.
The research team generated a high-quality draft genome of the genetic lineage H13 isolated from Hong Kong and compared it to the distantly related T. adhaerens. Substantial structural differences between the two genomes were generated, that point to a deep genomic separation and provide the support that adaptation by gene duplication is likely a crucial mechanism in placozoan speciation. We further provide genetic evidence for reproductively isolated species and suggest a genus-level difference of H13 to T. adhaerens, justifying the designation of H13 as a new species, Hoilungia hongkongensis nov. gen., nov. spec., now the second described placozoan species and the first in a new genus. The multilevel comparative genomics approach is, therefore, likely to prove valuable for species distinctions in other cryptic microscopic animal groups that lack diagnostic morphological characters, such as some nematodes, copepods, rotifers, or mites.
This study uses a comparative genomics approach for a deeper appreciation of the structure and causes of the deeply diverging lineages in the Placozoa. Placozoans are a phylum of tiny (approximately 1 mm) marine animals that are found worldwide in temperate and tropical waters. They are characterized by morphological simplicity, with only a handful of cell types, no neurons, no tissue organization, and even no axial polarity.
The research team generated a high-quality draft genome of the genetic lineage H13 isolated from Hong Kong and compared it to the distantly related T. adhaerens. Substantial structural differences between the two genomes were generated, that point to a deep genomic separation and provide the support that adaptation by gene duplication is likely a crucial mechanism in placozoan speciation. We further provide genetic evidence for reproductively isolated species and suggest a genus-level difference of H13 to T. adhaerens, justifying the designation of H13 as a new species, Hoilungia hongkongensis nov. gen., nov. spec., now the second described placozoan species and the first in a new genus. The multilevel comparative genomics approach is, therefore, likely to prove valuable for species distinctions in other cryptic microscopic animal groups that lack diagnostic morphological characters, such as some nematodes, copepods, rotifers, or mites.
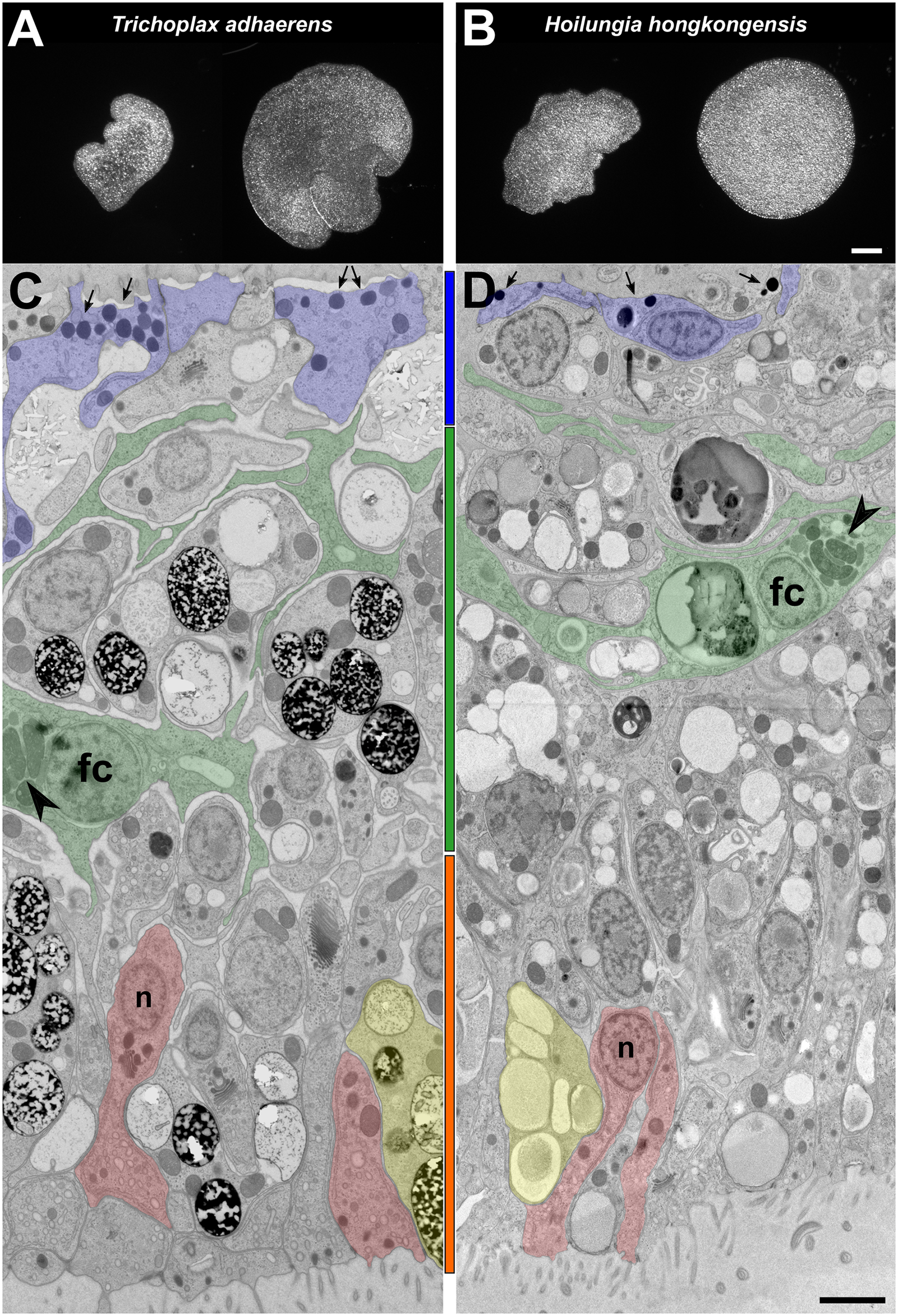
Comparative morphology of T. adhaerens and H. hongkongensis nov. gen., nov. spec.
This paper was written by ESR Project 1: Phylogenomics of non-bilaterian animals. This team works on the phylogenetic relationships among non-bilaterian animals. The robust resolution of these animals is necessary to understand early animal evolution and the emergence of key bilaterian traits, such as nervous systems, muscles, and guts. More non-bilaterian taxon sampling, especially from undersampled lineages within Porifera and Placozoa, as well as new primary sequence-independent phylogenetic markers, such as “rare genomic changes” (RGCs), are needed, in order to test the many controversially discussed hypotheses.
Original source: Eitel M, Francis WR, Varoqueaux F, Daraspe J, Osigus H-J, Krebs S, et al. (2018) Comparative genomics and the nature of placozoan species. PLoS Biol 16(7): e2005359. https://doi.org/10.1371/journal.pbio.2005359