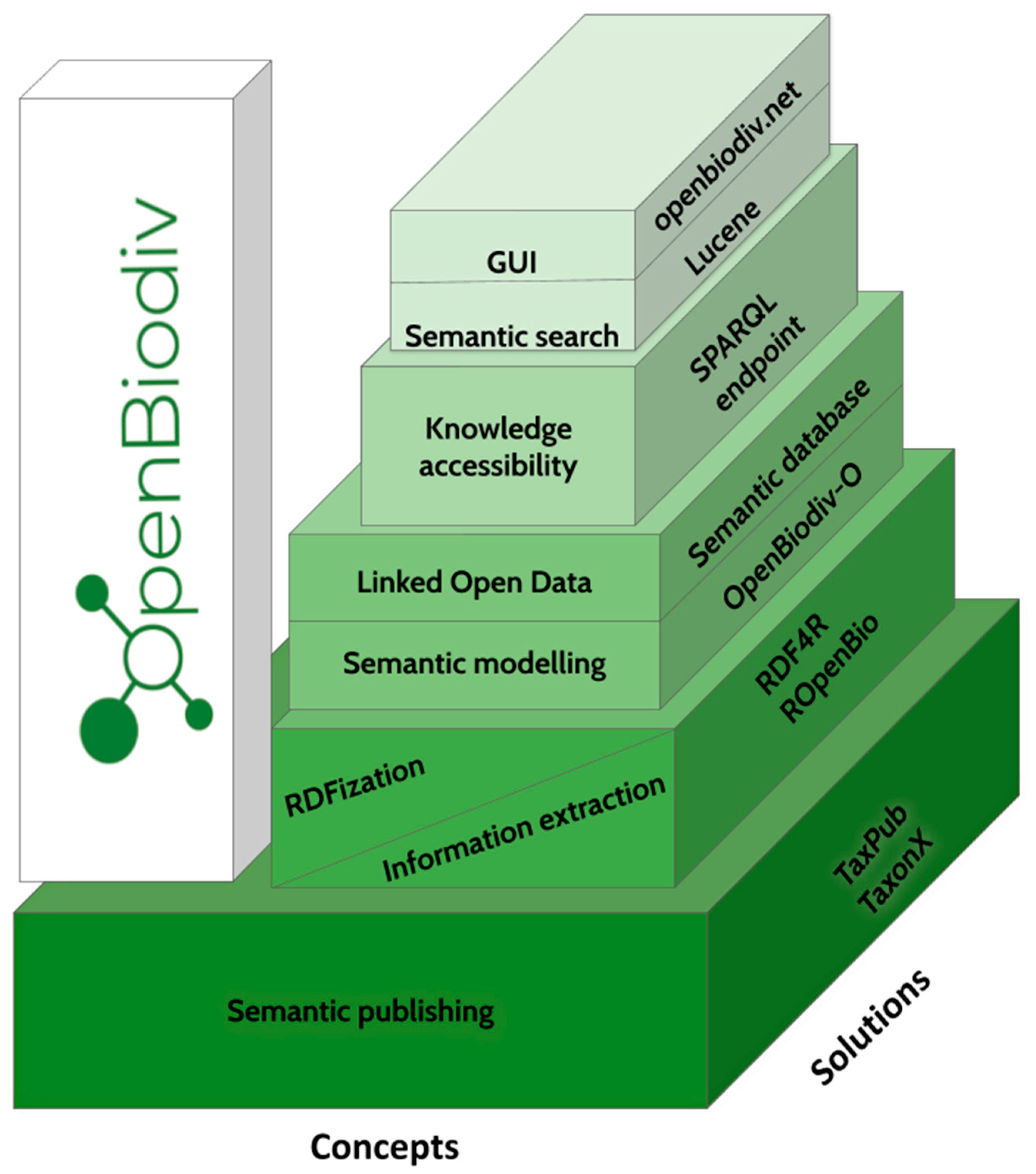
A new approach towards bridging the gap between biodiversity data and published narrative has been developed within the framework of IGNITE. With the application of semantic technologies, the novel system OpenBioDiv provides a long-anticipated mechanism for storage and management of diverse information of particular importance for biology.
Hundreds of years of biodiversity research have resulted in the accumulation of a substantial pool of communal knowledge; however, most of it is stored in silos isolated from each other, such as published articles or monographs. The need for a system to store and manage collective biodiversity knowledge in a community-agreed and interoperable open format has evolved into the concept of the Open Biodiversity Knowledge Management System (OBKMS). This paper presents OpenBiodiv: An OBKMS that utilizes semantic publishing workflows, text and data mining, common standards, ontology modelling and graph database technologies to establish a robust infrastructure for managing biodiversity knowledge. It is presented as a Linked Open Dataset generated from scientific literature.OpenBiodiv encompasses data extracted from more than 5000 scholarly articles published by Pensoft and many more taxonomic treatments extracted by Plazi from journals of other publishers. The data from both sources are converted to Resource Description Framework (RDF) and integrated in a graph database using the OpenBiodiv-O ontology and an RDF version of the Global Biodiversity Information Facility (GBIF) taxonomic backbone. Through the application of semantic technologies, the project showcases the value of open publishing of Findable, Accessible, Interoperable, Reusable (FAIR) data towards the establishment of open science practices in the biodiversity domain.