IGNITE researchers, Paschalis Natsidis (ESR 12) and Prof. Max Telford, have recently conducted a study on the Systematic errors in inference and their effects on evolutionary analyses, made available in the iScience Journal. The publication has its main focus on investigating the errors in identifying orthologous genes and has found that they increase with higher rates of evolution.
Orthology is a type of homology where the homologous genes originated at a speciation event. The evolution of orthologous genes and the fact that their relationships coincide with species phylogeny have made them key markers in evolutionary biology. The paper states that the availability of complete sets of genes from many organisms makes it possible to identify genes unique to (or lost from) certain clades. Using this data, it is possible to reconstruct phylogenetic trees, identify genes and in terms of phylostratigraphy, to recognise ages of genes in a certain species.
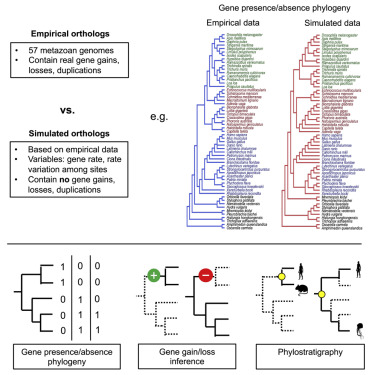
The study shows that there are errors present in the downstream analyses, pointed out by the researchers to be rather frequent and randomly distributed.
It is demonstrated that each of the results derived from simulated data are able to mirror observations from empirical information. Although some of the evolutionary signals found in the sets of orthologs derived from empirical data must represent real events of gene gain and loss, the results suggest that this signal is likely to be supplemented to an unknown degree by the systematic errors described.
The full paper you can read here.