A research article recently published in the Genome Biology journal presents a new software for genome scaffolding on a chromosome level. The paper called "InstaGRAAL: chromosome-level quality scaffolding of genomes using a proximity ligation-based scaffolder" outlines the development and testing of the InstaGRAAL software for large genome assembly based on chromosomal conformation capture data.
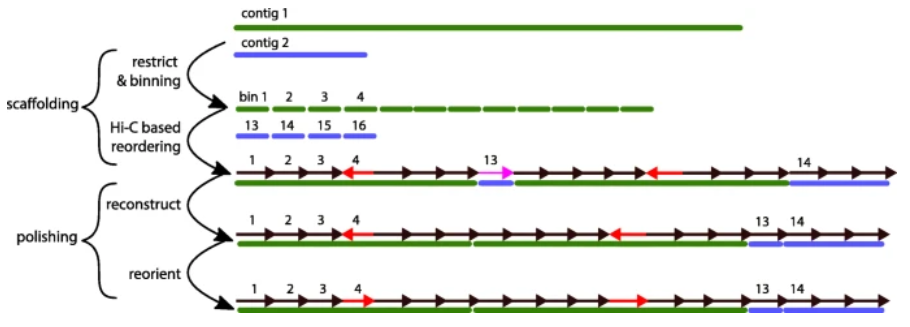
A step-by-step top to bottom correction procedure in InstaGRAAL. Source: Baudry, L., Guiglielmoni, N., Marie-Nelly, H. et al. instaGRAAL: chromosome-level quality scaffolding of genomes using a proximity ligation-based scaffolder. Genome Biol 21, 148 (2020). https://doi.org/10.1186/s13059-020-02041-z
The authors of the article, amongst whom the IGNITE researcher Nadège Guiglielmoni, contributed towards the development of the instaGRAAL software as a complete overhaul of the GRAAL program, which was adapted to allow efficient assembly of large genomes. The new instaGRAAL software offers a number of improvements of GRAAL, including a modular correction approach which allows for the integration of independent data.
To perform genome scaffolding, the instaGRAAL tool uses chromosome conformation (Hi-C) data. The research team that developed the tool validated the program’s functioning by generating near-complete genome assemblies for two brown algae species, and humans, with minimal human intervention.
Read the article here.