IGNITE researchers Dr. Nadège Guiglielmoni and Prof. Jean-Francois Flot have recently published a research article titled “Overcoming uncollapsed haplotypes in long-read assemblies of non-model organisms”. The paper, which was published in the open access journal BMC Bioinformatics, provides a thorough evaluation of popular assemblers on a non-model eukaryote genome with variable levels of heterozygosity. It concludes that PacBio and Nanopore technologies become more accessible in technicity and in cost, and therefore long-read assemblers flourish and are starting to deliver chromosome-level assemblies.
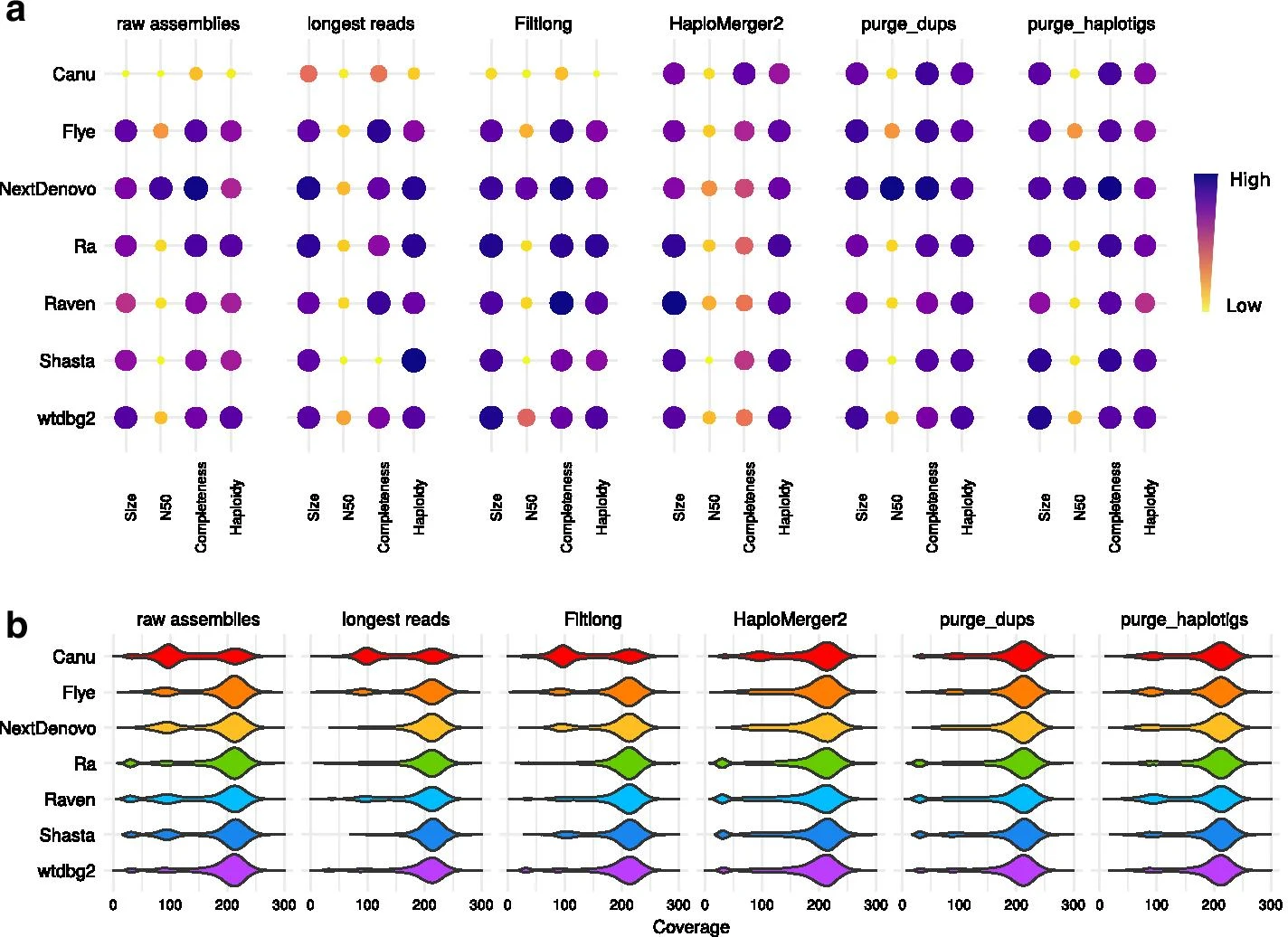
Figure 1: Statistics of PacBio assemblies. Statistics of raw assemblies obtained from the full PacBio dataset (raw assemblies), with a preliminary read-filtering step (keeping only reads larger than 15 kb, or those selected by Filtlong based on quality and length), or a subsequent removal of uncollapsed haplotypes with HaploMerger2, purge_dups, or purge_haplotigs. a Assembly scores for size, N50, completeness and haploidy. b Long-read coverage distribution over the contigs.
The authors of the article tested different assembly strategies of the rotifer Adineta vaga, a non-model organism for which high coverages of both PacBio and Nanopore reads were available. The assemblers that they tested (Canu, Flye, NextDenovo, Ra, Raven, Shasta and wtdbg2) exhibited strikingly different behaviors when dealing with highly heterozygous regions, resulting in variable amounts of uncollapsed haplotypes.
You can read the full publication here.