A research article, co-authored by IGNITE members Mariya Dimitrova and Prof. Lyubomir Penev and published in the open access journal GigaScience, presents a template and workflow for automatic import of standard-compliant metadata into an omics data paper manuscript within a journal publishing system. These advances provide a mechanism for enhancing existing metadata through publishing.
The researchers aimed to establish a new type of article structure and template for omics studies: the omics data paper. To improve data interoperability and further incentivize researchers to publish well described datasets, the scientists created a prototype workflow for streamlined import of genomics metadata from the European Nucleotide Archive directly into a data paper manuscript.
The researchers aimed to establish a new type of article structure and template for omics studies: the omics data paper. To improve data interoperability and further incentivize researchers to publish well described datasets, the scientists created a prototype workflow for streamlined import of genomics metadata from the European Nucleotide Archive directly into a data paper manuscript.
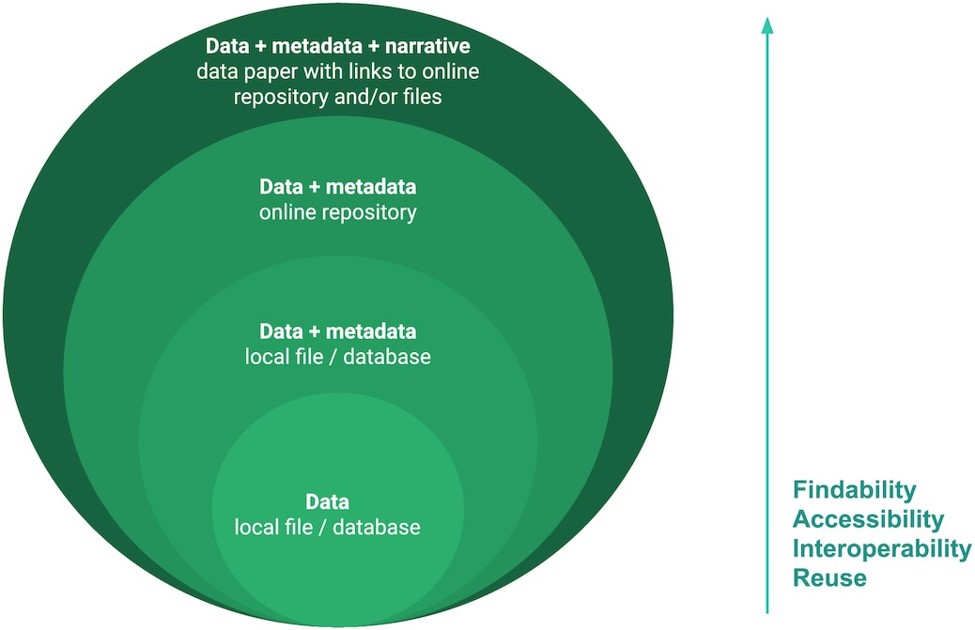
Figure 1: The different layers of FAIRness of data and metadata. Describing data and metadata in a data paper.
Promoting enhanced metadata descriptions and enforcing manuscript peer review and data auditing of the underlying datasets brings additional quality to datasets. The researchers hope that streamlined metadata reuse for scholarly publishing encourages authors to create enhanced metadata descriptions in the form of data papers to improve both the quality of their metadata and its findability and accessibility.
You can read the full publication, titled “A streamlined workflow for conversion, peer review, and publication of genomics metadata as omics data papers”, here.