Supervisor: Jean-François Flot (University of Brussels), Co-supervisor Romain Koszul (Institut Pasteur, Paris)
Student: Nadège Guiglielmoni
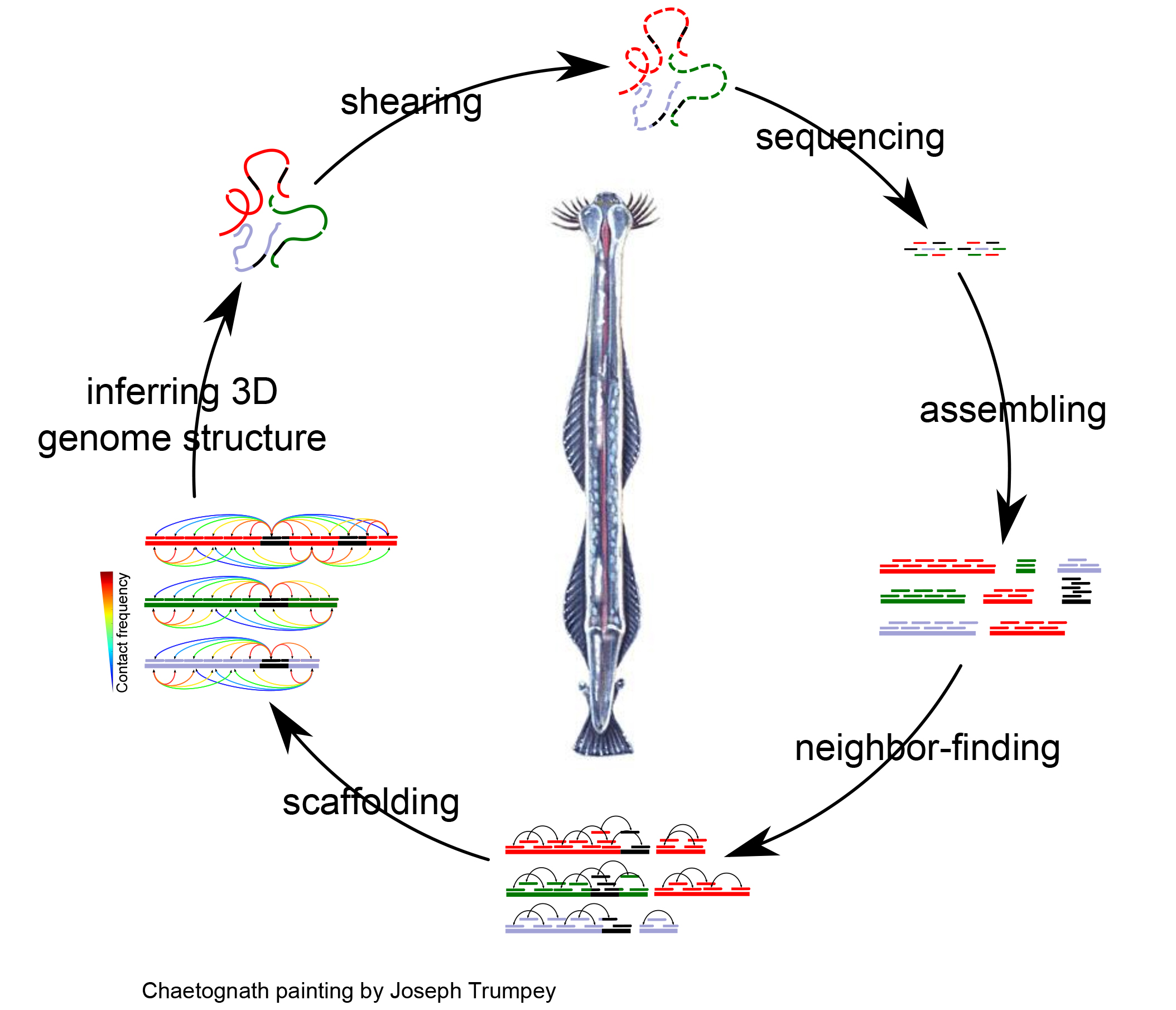
Objectives: Despite a near-exponential increase in the number of eukaryotic genome projects over the last few year, the vast majority of them yield only "permanent drafts", i.e., genomes that remain heavily fragmented at the end of the project and are deposited as such in public databases. This is because the large and abundant repeats found in eukaryotic genomes make them extremely difficult to bring to the "golden standard" of one contig per chromosome. Bleeding-edge chromosome conformation capture (3C) approaches, however, appear as a promising path to overcome the repeat issue and could make "perfect" genome assemblies a routine outcome for eukaryotic genomes, thereby increasing the power of downstream analyses.
O1: Improve existing 3C procedures to make them more suited to non-model invertebrates.
O2: Improve existing computational pipeline to enable de novo assembly of 3C data from invertebrates with maximal accuracy and completeness.
O3: Apply these approaches to the genome of a chaetognath (a group whose genome has proved very challenging to assemble using conventional approaches) and shed light on its evolution by comparative genomics.
Secondments: Romain Koszul (Institut Pasteur, Paris), Eduardo Pareja (Granada), Yu Wang (Leibniz-Supercomputing Centre of the Bavarian Academy of Science and Humanities, Munich).